Research
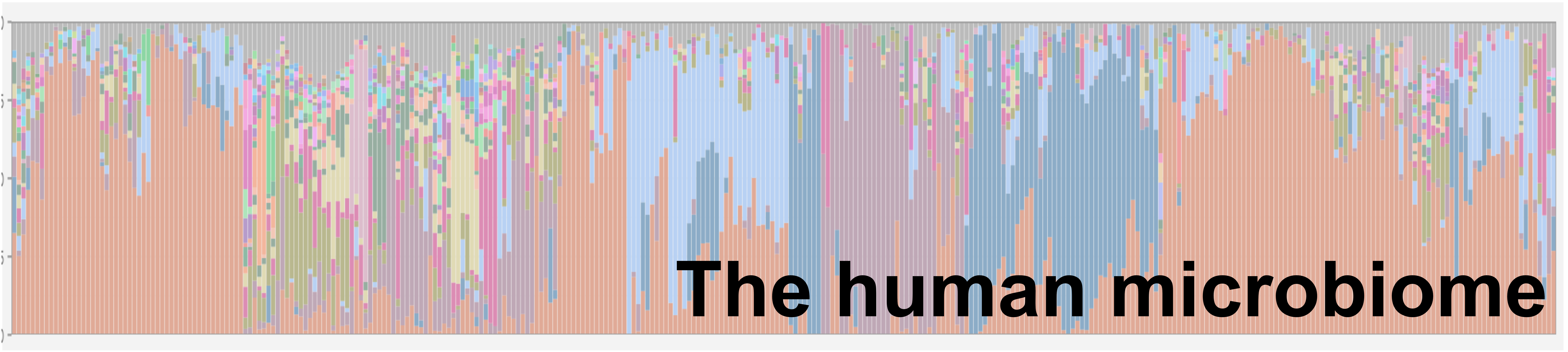
The microbes that live on and in our bodies – has been implicated in a myriad of diseases. The human microbiome generally consists of commensal organisms with beneficial behaviours (e.g. gastrointestinal digestion, immune maturation). However, dysbiosis of the human microbiota has been implicated in many diseases/disorders (IBD, IBS, obesity, cancers) with some evidence for a role in others (autism, depression). Often, dysbiosis is described as a difference in the overall community structure; however, to mitigate these diseases, we must understand the key microbial drivers. However, because this field is still relatively new, we don’t have the algorithms and software to properly understand these communities. The focus of the lab is to develop, test, and use a new computational methodology to better understand the microbiome. We then use these tools in conjunction with high-throughput culture-enrichment to test hypotheses about the role of the human microbiome in disease.
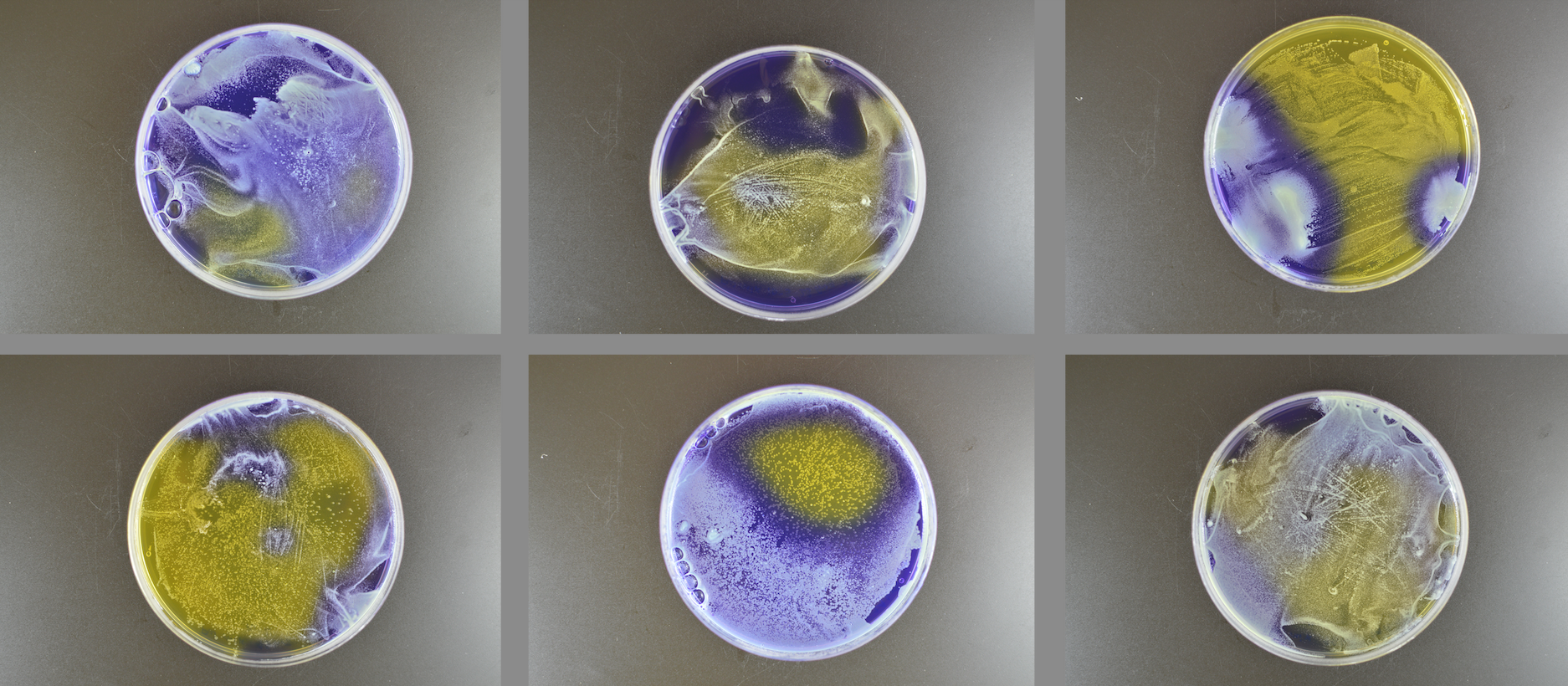
Microbes co-exist within microbiomes. They can compete with each other via a variety of mechanisms including antimicrobials and bacteriocins. Our lab and others have found that these interactions are strain specific, meaning that some strains of a particular microbe may induce competition in another strain, but others do not. We are particularly interested in how bacterial concentration, growth medium, and the environment affect these relationships.

Genes are rarely used in isolation but instead together form multi-protein complexes and are part of functional pathways. However, we still have only annotated a fraction of the diversity of genes present across bacterial life. We think that one way to help better characterize this genetic diversity is by interrogating gene-gene association/co-occurrence patterns across large sets of genomes and/or metagenomes.

Pangenomes describe the genetic diversity within a given set of organisms (for e.g., strains of a species). The genes within a pangenome can either be "core" - present in all or most strains - or "accessory" - present in some strains. Accessory genes can include genes which as those involved in anti-microbial resistance (AMR). In the lab, we are interested in understanding gene-gene association patterns that exist across groups of strains.
Research in the Whelan lab is funded by:























